Zadanie Inżynieria genetyczna (inz)
Pomóż nam usprawnić bazę zadań!
Genetic Engineering
Memory limit: 128 MB
Byteotian paleoarchaeologists recently unearthed a few ambers, which had trapped ancient mosquitoes inside. After analysing the samples of insects it turned out that they come from the Jurassic period, and therefore likely to have been in contact with large reptiles that dominated the Byteotian lands. This gave geneticists a quaint idea: to try to recover byteoraptor genetic material from the blood of mosquitoes.
Byteoraptor genome, as in all Bytean organisms, is a chain consisting of a number of byteo-aminoacids. For simplicity we denote the types of byteo-aminoacids by natural numbers. Redundancy occurs in a genome-every type of byteo-aminoacid is repeated 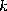 times (specifically, the length of each correct genome is a multiple of
times (specifically, the length of each correct genome is a multiple of 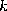 ). In other words, if we divide the genome into blocks consisting of subsequent
). In other words, if we divide the genome into blocks consisting of subsequent 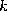 byteo-aminoacids, each block will contain byteo-aminoacids of the same kind.
byteo-aminoacids, each block will contain byteo-aminoacids of the same kind.
Geneticists were able to isolate a suspected chain consisting of byteo-aminoacids, from the blood of a mosquito, being  in length. Unfortunately, the chain may not be a valid genome-scientists suspect that it may have been contaminated by foreign byteo-aminoacids. Presently they want to test their hypothesis and remove the least byteo-aminoacids from that chain, such that a normal genome emerges. In case of many equally good possibilities, the researchers are interested in the genome that is the earliest in lexicographical order*. Your task is to help them to make a breakthrough discovery.
in length. Unfortunately, the chain may not be a valid genome-scientists suspect that it may have been contaminated by foreign byteo-aminoacids. Presently they want to test their hypothesis and remove the least byteo-aminoacids from that chain, such that a normal genome emerges. In case of many equally good possibilities, the researchers are interested in the genome that is the earliest in lexicographical order*. Your task is to help them to make a breakthrough discovery.
*Let 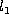 and
and 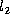 be two different chains of the same length, consisting of byteo-aminoacids. To determine which one is earlier in lexicographical order it is necessary to find the first position where the chains differ. The chain earlier in the lexicographical order is the one which has byteo-aminoacid marked with a lower number in this position.
be two different chains of the same length, consisting of byteo-aminoacids. To determine which one is earlier in lexicographical order it is necessary to find the first position where the chains differ. The chain earlier in the lexicographical order is the one which has byteo-aminoacid marked with a lower number in this position.
Input
The first line contains two integers
 and
and 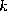 (
(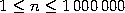 ,
, 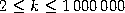 ): the length of extracted chain of byteo-aminoacids and redundancy degree of a correct genome. The second line contains a sequence
): the length of extracted chain of byteo-aminoacids and redundancy degree of a correct genome. The second line contains a sequence  of integers
of integers
 (
(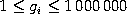 ): the types of subsequent byteo-aminoacids in the chain.
): the types of subsequent byteo-aminoacids in the chain.
Output
The output should contain two lines. The first one should contain the number 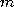 (
(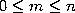 ) denoting length of the longest proper genome, which may arise by removing some byteo-aminoacids from the specified chain.
) denoting length of the longest proper genome, which may arise by removing some byteo-aminoacids from the specified chain.
The second line should contain a chain of 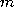 numbers describing the types of subsequent byteo-aminoacids in the correct genome. In case there are multiple solutions, your program should output the smallest lexicographically. If
numbers describing the types of subsequent byteo-aminoacids in the correct genome. In case there are multiple solutions, your program should output the smallest lexicographically. If 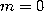 (i.e. geneticists have failed to isolate any non-empty correct genome), the second line of output should be empty.
(i.e. geneticists have failed to isolate any non-empty correct genome), the second line of output should be empty.
Example
For the input data:
16 3 3 2 3 1 3 1 1 2 4 2 1 1 2 2 2 2
the correct result is:
9 1 1 1 2 2 2 2 2 2
Task author: Tomasz Idziaszek
Kontakt
In the event of technical difficulties with Szkopuł, please contact us via email at [email protected].
If you would like to talk about tasks, solutions or technical problems, please visit our Discord servers. They are moderated by the community, but members of the support team are also active there.

 English
English